Fig. 1.
From: Enhanced transcriptome-wide RNA G-quadruplex sequencing for low RNA input samples with rG4-seq 2.0
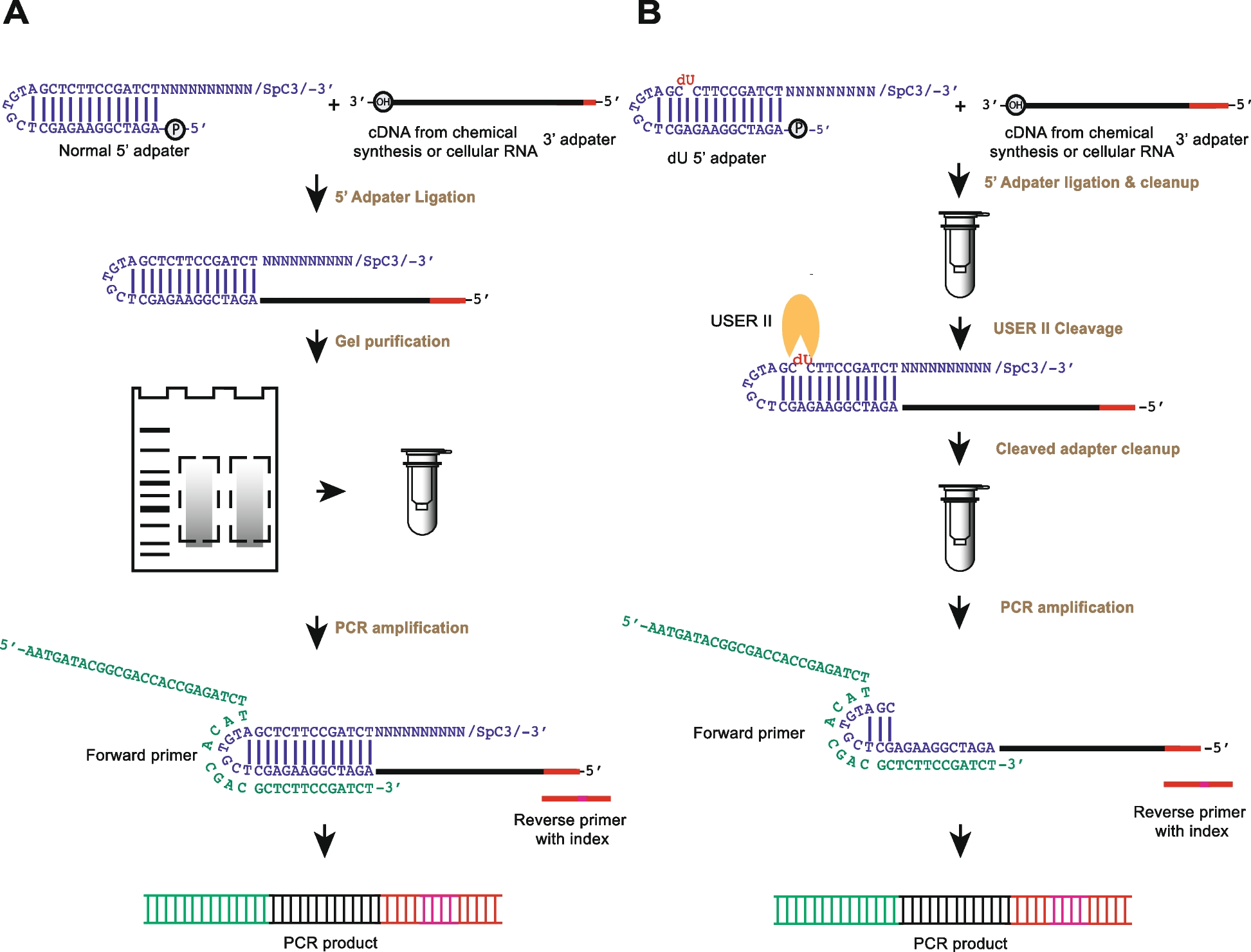
Comparison of the experimental strategy of rG4-seq and rG4-seq 2.0 from 5‘ adapter ligation to PCR amplification. The sequences of the oligonucleotides used can be found in Additional file 1: Table S1. The experimental pipelines that use A normal 5′ adapter and B 5′ dU adapter are shown (blue). In A, the normal 5′ adapter ligates to the cDNA (black) with 3′ adapter sequence (red) followed by gel purification before performing PCR amplification. The 6nt index region in the reverse PCR primer is in pink. In B, dU adapter is used for the ligation, and after the column purification, the enzymatic cleavage step is performed with cleavage enzyme (USER II, orange pacman) followed by column clean-up before the PCR amplification. USER II enzyme cleaves the 5′ adapter at the dU position as indicated in red in the 5′ dU adapter. The dU adapter contains the same secondary structure as the normal 5′ adapter and carries a 5′ phosphate at its 5′ end to ligate to incoming cDNA with a 3′hydroxyl, and a C3 spacer group at its 3′ end to block and avoid self-oligomerization. The 10 random nucleotides (N10) serve as a template for the hybridization of any incoming cDNA
