Fig. 1
From: Introgressions lead to reference bias in wheat RNA-seq analysis
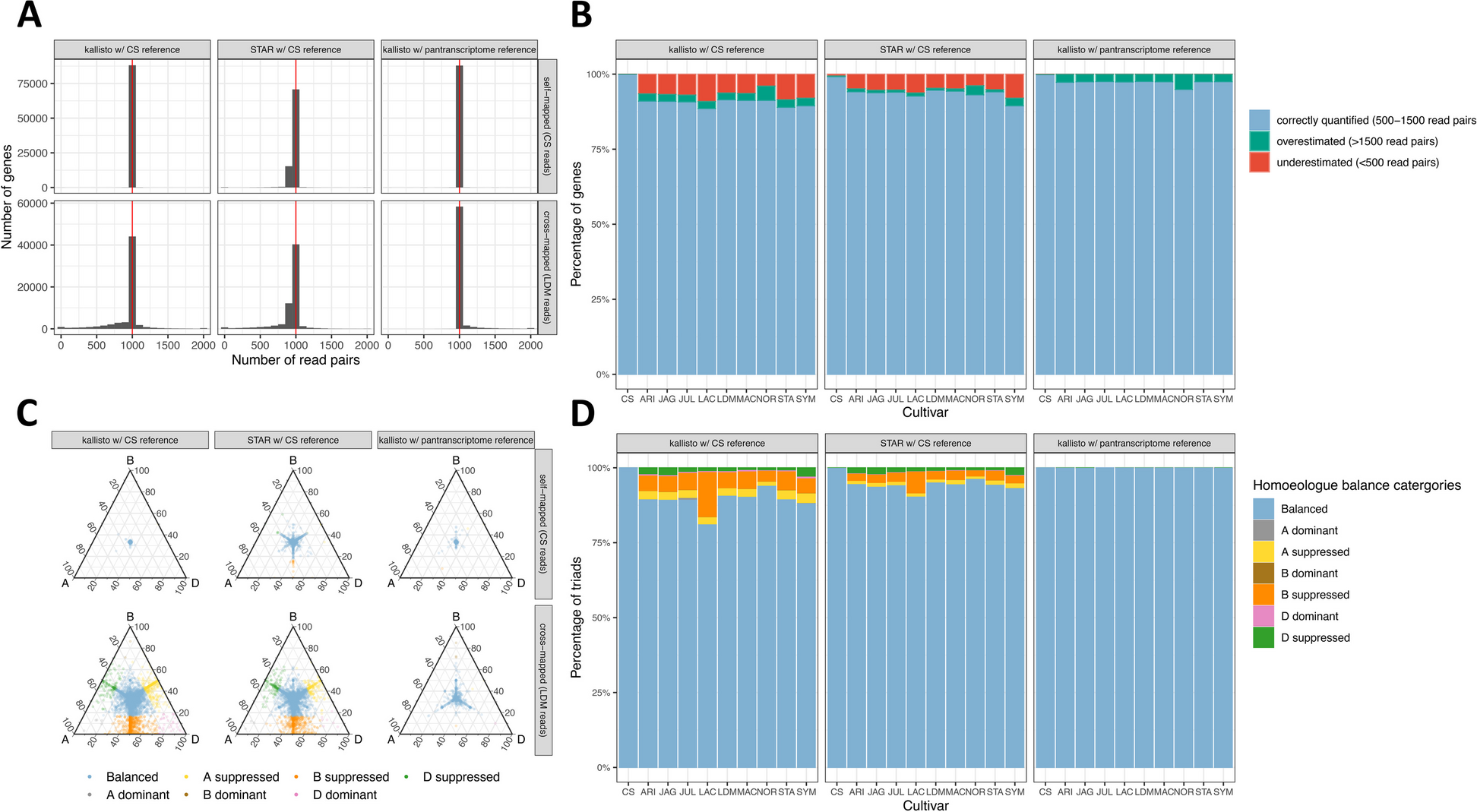
Assessing the extent of reference bias in wheat. A Distribution of read counts when self-mapping Chinese Spring simulated reads or cross-mapping Landmark simulated reads. Comparing STAR and kallisto using the Chinese Spring RefSeq v1.0 reference and RefSeq v1.1 transcriptome and kallisto using the pantranscriptome reference. B Percentage of genes with expression estimated correctly, expression underestimated (< 500 read pairs) and expression overestimated (> 1500 read pairs) for simulated reads from 10 cultivars aligned to Chinese Spring with kallisto and STAR or to the pantranscriptome reference with kallisto. C Balance of homoeologue expression across triads when self-mapping Chinese Spring or cross-mapping Landmark simulated reads, comparing STAR and kallisto using the Chinese Spring RefSeq v1.0 reference and RefSeq v1.1 transcriptome and kallisto using the pantranscriptome reference. Each point on the ternary plot represents one triad. Points towards a corner indicate dominant expression of that homoeologue, and points opposite a corner indicate suppression of that homoeologue. D Percentage of triads in each expression category, using simulated reads from 10 cultivars aligned to Chinese Spring with kallisto and STAR or to the pantranscriptome reference with kallisto
